Scientific success stories based on Biocenter Finland technology services
Biobank technologies
Bioinformatics
Drug discovery & chemical biology
Electron microscopy
Genome-wide methods
Mouse models
Non-mammalian model organisms
Metabolomics
Protein proteome
Real-time imaging
Single-cell omics
Stem cells & genome editing
Structural biology
Viral gene transfer & cell therapy
See more on Biocenter Finland's achievements and impact in the Annual Report
Biobank technologies
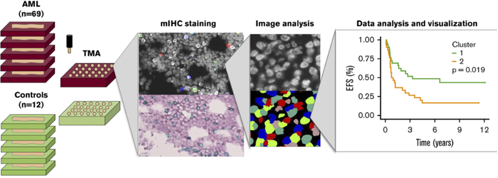
Bruck and others (2020) analysed thirty immunomarkers using multiplexed immunohistochemistry (mIHC) and computerized image analysis at single cell level. Quantitative immunophenotyping distinguished different classes of leukemia (AML, CML, B-ALL) but also revealed distinct prognostic subgroups (Fig 1). Especially, a higher proportion of T regulatory cells (Tregs) predicted inferior survival of patients with acute myeloid leukemia (AML). The results suggest a rationale for including immunologic parameters to improve disease classification and patient risk stratification in AML.
Figure. Flow diagram of the study on multiplexed immunophenotyping combined with image analysis for classification of patients with leukemia
Bioinformatics
Academician Sirpa Jalkanen discovered a previously undefined role for medullary lymphatic endothelial cells in human immunity. Her findings were facilitated by the BF Genomic-wide methods and BF Bioinformatics Platforms and provide valuable new information with potential to help improving human health and prevent diseases. (Takeda et al. Single-Cell Survey of Human Lymphatics Unveils Marked Endothelial Cell Heterogeneity and Mechanisms of Homing for Neutrophils. Immunity 51(3):561-572.e5, 2019.
Professor Mika Rämet discovered a new molecular component of the signaling network that promotes spontaneous preterm birth, the leading cause of neonatal death and morbidity worldwide. His finding was facilitated by the support provided by the BF Genome-wide methods and BF Bioinformatics Platforms. (Tiensuu et al. Risk of spontaneous preterm birth and fetal growth associates with fetal SLIT2. PLoS Genetics 15(6):e1008107, 2019.
Professor Anna-Liisa Levonen discovered a non-classical activation mechanism in the lethal brain cancer, glioma, providing a rationale to target the Nrf2-regulatory pathway in a subset of patients. The implications of this study on cancer diagnostics are in promoting the wider adoption of multi-omics characterization of patient samples, as the mechanism for oncogenic activity would have been missed by DNA mutation data alone. Data analyses were facilitated by the BF Bioinformatics Platform. (Pölönen et al. Nrf2 and SQSTM1/p62 jointly contribute to mesenchymal transition and invasion in glioblastoma. Oncogene 38(50):7473-7490, 2019.
Drug discovery and chemical biology
As a part of long-lasting collaboration between BCK-PCM and University of North-Carolina at Chapel Hill, a systematic study to develop new and specific kinase inhibitors against selected kinases has continued. As an example of results, we were able to develop quantitative structure-activity relationship models and further optimized a series of very selective GAK/SLK/STK10 kinase inhibitors. Some of the key points have been recently published. (1)
Another recent example of BCK-PMC services identification and optimization of CK2 kinase inhibitors together with Prof. Joachmi Jose and his group at University of Münster. A natural ligand, Bikaverin, was identified via virtual screening (using a publicly available ZINC database and CSC taito supercomputer) and this hit compound has now been used as a starting point for a more systematic CK2 inhibitor development project. (2)
(1) Asquith, C. R. M.; Tizzard, G. J.; Bennett, J. M.; Wells, C. I.; Elkins, J. M.; Willson, T. M. M.; Poso, A.; Laitinen, T. Targeting the Water Network in Cyclin G Associated Kinase (GAK) with 4-Anilino-Quin(Az)Oline Inhibitors. Biorxiv 2020, 2020.03.06.976563. https://doi.org/10.1101/2020.03.06.976563.
(2) Haidar, S.; Aichele, D.; Birus, R.; Hielscher, J.; Laitinen, T.; Poso, A.; Jose, J. In Vitro and in Silico Evaluation of Bikaverin as a Potent Inhibitor of Human Protein Kinase CK2. Molecules (Basel, Switzerland)2019, 24 (7). https://doi.org/10.3390/molecules24071380.
Electron microscopy
UEF-EM participated in a few studies related to pharmaceuticals and drug development. Hellinen et al. published a study in Scientific Reports using ARPE-19 cells internalized with melanosomes to study drug uptake. Melanin pigment has a significant role in ocular pharmacokinetics, because many drugs bind at high extent to melanin in the retinal pigment epithelial cells.
Hellinen L, Hagström M, Knuutila H, Ruponen M, Urtti A, Reinisalo M. Characterization of artificially re-pigmented ARPE-19 retinal pigment epithelial cell model. Sci Rep. 2019 Sep 24;9(1):13761. doi: 10.1038/s41598-019-50324-8.
Genome-wide methods
A breakthrough study by Prof Sirpa Jalkanen and collaborators examines the role of lymphatic vessels with focus on lymphatic endothelial cells (LEC). The group profiled 33 000 cells and identified 6 LEC subtypes. The study provides comprehensive characterization of LEC heterogeneity and reveals a previously undefined role for medullary LECs in human immunity with implications to cancer metastasis. This study was a cross-platform collaboration between GWM, Single-Cell Omics and Bioinformatics platforms, with GWM providing expertise for library preparation and next-generation sequencing service for single cell RNA sequencing.
(Takeda A et al. Single-Cell Survey of Human Lymphatics Unveils Marked Endothelial Cell Heterogeneity and Mechanisms of Homing for Neutrophils. Immunity. 2019 Sep 17;51(3):561-572).
Light microscopy
As imaging is very critical in life science research, the platform has continued to have a central role in several scientific breakthroughs and in solving significant research problems. Some concrete examples:
- Modern laser scanning confocal microscopy at CIC-TBI played a crucial role in Academician Sirpa Jalkanen’s research to reveal the structures of lymphatic tissues, unveiling previously undefined functions in human immunity (Single-Cell Survey of Human Lymphatics Unveils Marked Endothelial Cell Heterogeneity and Mechanisms of Homing for Neutrophils. Takeda A. et al. 2019. Immunity, 51: 561-572.)
- FIMM-HCA developed a new software tool for image registration and quantification in spatial tissue analysis, successfully used to analyse oncogenic signalling in lung cancer. (Spa-RQ: An Image Analysis Tool to Visualise and Quantify Spatial Phenotypes Applied to Non-Small Cell Lung Cancer. Bao et al. 2019. Sci Rep, 9: 17613.)
- Several microscopes from BIU-LM, including superresolution imaging, were central in Prof. Elina Ikonen’s group’s work in developing an improved version of a powerful tool to study protein function. This work benefits scientists from all domains of cell biological research. (An efficient auxin-inducible degron system with low basal degradation in human cells. Li et al. 2019. Nat Methods, 16: 866-869.)
- The lab of Ari-Pekka Mähönen utilized LMU-BI microscopes to study how plant growth originates, confirming a significant classical theory of plant growth (Sanio 1873). (High levels of auxin signalling define the stem-cell organizer of the vascular cambium. Smetana et al. 2019. Nature, 565: 485-489.)
- A wide range of advanced imaging tools at TIC-BCO played a crucial role in a project where the role of collagen XIII protein in bone remodelling and osteoporosis was characterized. (Collagen XIII-derived ectodomain regulates bone angiogenesis and intracortical remodeling. Koivunen et al. 2019. Matrix Biol, 83: 6-25.)
Mouse models
Altogether, several hundred scientists benefit from the FinGMice services in Finland annually. Importantly, the rodent models are essential tools to many of the leading Finnish medical and biomedical research groups led by Academians, Academy professors, and professors at Centres of Excellence of Academy of Finland.
GRACILE syndrome is a severe neonatal metabolic disease of the Finnish disease heritage caused by a point mutation in the nuclear BCS1L gene. A mouse model that recapitulates human GRACILE syndrome has been extensively studied in Kallijärvi-Fellman research group at the Folkhälsan Research Center and the University of Helsinki with the help of national and international mouse phenotyping services. Their recent study on GRACILE mouse model, published in the journal Nature Communications, further facilitates the development of diagnostic, prognostic and therapeutic strategies for patients suffering this devastating lethal metabolic disease in neonates and provides a new tool for studies of mitochondrial diseases.
Purhonen J, Grigorjev V, Ekiert R, Aho N, Rajendran J, Pietras R, Truvé K, Wikström M, Sharma V, Osyczka A, Fellman V, Kallijärvi J A Spontaneous Mitonuclear Epistasis Converging on Rieske Fe-S Protein Exacerbates Complex III Deficiency in Mice. Nat Commun. 2020 Jan 16;11(1):322.
Non-mammalian model organisms
In 2019, the group of Osamu Shimmi (Institute of Biotechnology, Helsinki) published a landmark paper in PNAS, describing a major discovery in the field of morphogenesis. Using ModOrgNon's Drosophila services, the group demonstrated how changes in tissue shape in 3D affect signaling by morphogens, diffusible substances which establish the specific fates of different cell-types during development, in a gradient fashion. Morphogens not only affect tissue shape, but there is also a feedback mechanism from the 3D architecture to the morphogen gradient itself.
Metabolomics
FIMM-Meta has increased its local and international academic and clinical collaborations, and the Unit had 23 projects started or ongoing during 2019. One local academic project with Prof. Anu Wartiovaara (Suomalainen) from University of Helsinki and another one with Prof. Del Chiaro from Karolinska Institute were both published in the high impact journal Scientific Reports, FIMM PI as co-author. The targeted 100 metabolite method (method published 2018 by Velagapudi) was successfully used in both articles. In addition, targeted SCIEX lipidyzer 1000 lipid was combined with the 100 metabolite method to create integrated metabolomics and lipidomics approach.
ViMU was involved as a co-author in peer-reviewed scientific publications where metabolomics and/or mass spec data was provided by the unit. A list of publications linked to ViMU can be found from the Tuhat-database (infrastructures) of University of Helsinki (http://www.helsinki.fi/tuhat/, search for ViMU).
The work at BCK has continued on metabolomics applications for food, health, toxicology, and nutrition studies. The specific topics include the effects of environmental pollutants and alcohol on the offspring or on nutrition effects of nutrition in disease. BCK has been involved in 48 projects in the above-mentioned areas of research. In year 2019 BCK had analyzed over 42,000 samples (20,676 and 22,134 samples in HRMS and QQQ instruments, respectively) involved in these projects. These research results will be and are published in highly reputed international journals. A major deal of the publications is used in PhD theses. BCK was involved in 16 peer-reviewed scientific publications; as a co-author, where analytical services were provided by the unit.
Protein-proteome
CCAAT enhancer–binding protein epsilon (C/EBPε) is a transcription factor involved in late myeloid lineage differentiation and cellular function. The aim of this study was to molecularly characterize the effects of C/EBPε transcription factor Arg219His mutation identified in a Finnish family with previously genetically uncharacterized autoinflammatory and immunodeficiency syndrome. Genetic analysis, proteomics, genome-wide transcriptional profiling by means of RNA-sequencing, chromatin immunoprecipitation (ChIP) sequencing, and assessment of the inflammasome function of primary macrophages were performed. We describe a novel autoinflammatory disease with defective neutrophil function caused by a homozygous Arg219His mutation in the transcription factor C/EBPε. Mutated C/EBPε acts as a regulator of both the inflammasome and interferome, and the Arg219His mutation causes the first human monogenic neomorphic and noncanonical inflammasomopathy/ immunodeficiency. The mechanism, including widely dysregulated transcription, is likely not unique for C/EBPε. Similar multiomics approaches should also be used in studying other transcription factor–associated diseases.
Göös H, Fogarty CL, Sahu B, Plagnol V, Rajamäki K, Nurmi K, Liu X, Einarsdottir E, Jouppila A, Pettersson T, Vihinen H, Krjutskov K, Saavalainen P, Järvinen A, Muurinen M, Greco D, Scala G, Curtis J, Nordström D, Flaumenhaft R, Vaarala O, Kovanen PE, Keskitalo S, Ranki A, Kere J, Lehto M, Notarangelo LD, Nejentsev S, Eklund KK, Varjosalo *, Taipale J, Seppänen MRJ. J Allergy Clin Immunol. 2019
Real-time imaging
Demonstration of light-induced release of a cargo from nanoparticles was probably the most impressive research achievement of 2019 in the RTI unit. At the laboratories of Arto Urtti (Division of Pharmaceutical Biosciences, UH) and Dr Tatu Lajunen (Tokyo University) researchers have developed special nanoparticles engineered to deliver the cargo when they are shined with near infrared light. This smart nanodevice makes a drug accessible at the place it is needed, avoiding its dissemination throughout the body and the unwanted side effects most drugs have. This would increase considerably the amount of drug administered, since the dose will not be lost systemically. The nanoparticles were loaded with [99mTc]HMPAO and administered subcutaneously. Upon NIR irradiation, the nanoparticles disintegrate and the radioactive contents are released resulting in excretion of the radioactivity in urine. Additionally, the RTI unit conducted the evaluation of two novel drug delivery system scaffolds based on nanocrystalline cellulose and triple-PEGylated PSi using 111In SPECT with high impact especially for the former to a dual radio- and chemotherapeutic theranostic construct for B-Raf mutated metastatic melanoma (studies ongoing).
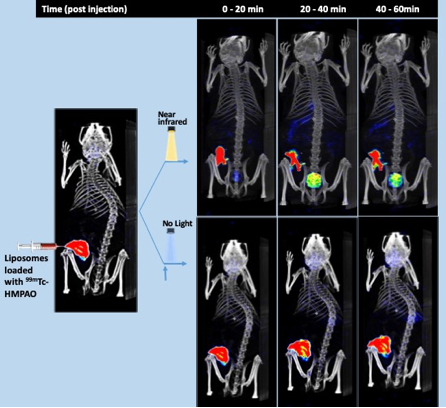
Fig. Time resolved SPECT/CT images of cargo (99mTc-HMPAO) release and distribution from NIR light sensitive liposomes in mice. Shown are representative images of two mice, after radiotracer loaded liposomes injection, and near infrared laser shone to one mice (top panel) but not the other (no light, bottom panel) at the area of injection. Around 4 MBq of 99mTc-hexamethyl-propyleneamine oxime (HMPAO), were injected in around 10-20 µl of saline. A total of 36 X SPECT images (every 10° from 360°) were recorded during the following 0 to 20, 20 to 40 and 40 to 60 min after injection. Shown are the SPECT signal average during the recording time in each image. The before light SOECT image is a reconstruction of the first 4 (orthogonal) projections (2-4 min) of the “no light” experiment, superimposed to the same CT anatomical reconstruction recorded at the time 120 min after injection. Control free tracer administrated mice showed similar pattern than top images but at slower kinetics (not shown).
Single cell omics
The last couple of years have seen groundbreaking advances of single cell technologies. Single cell analyses have allowed exploring complexity of tissues at highest resolution, description of development in tissues and organs, identification of cell types that contribute to human diseases, identification of unique cell types for organisms and studying organ evolution. Concrete examples are finding and analyzing rare circulating tumor cells (CTCs) from patients´ blood or studying drug resistant cell populations in cancer patients.
Biocenter Finland sc-omics platform is still a relatively novel infrastructure but it has already been realized that the impact of single cell technologies for Finnish science is going to be huge. In many levels of biomedical research from basic research to personalized medicine single cell analysis methods will be a very important feature of the entire palette of approaches used. This is also reflected in the diversity of user base that the sc omics platform already has.
Stem cells
In 2019 Stem cell platform partners published 33 new research results/technologies related to the iPSC and genome editing for genetic, pharmacology, artificial intelligence, and biomedical research. A concrete example of a solved research problem or a scientific breakthrough provided by each partner of the stem cell consortium is presented below.
HiLIFE: PMID: 31676859; 31161346; 30728358; 30270471. These publications highlight the advantage of using iPSC derived beta cells and genome editing for modeling pancreatic b-cell pathology and CRISPR activators technology to study cancer.
TAU PMID: 30837492, 31208058, 32079026, 31375001, 31163704. These publications highlight the use of iPS cells to study genetic regulators of hepatic differentiation, epigenetic memory of stem cell derived cells, and heart failure. The important technology development relates to the machine learning and organ-on-culture studies and their potential application in stem cell research.
BCK PMID: 31522977; 31477693; 31455857, 31016348, 30453390. Differentiation of hiPSC lines into fully functional brain microglia, astrocytes, cortical neurons, and endothelial cells that maintain disease pathology and can be used for transcriptomics, proteomics, and metabolomics analyses with a low number of lines. The studies also demonstrate the advantage of using iPSC-derived cells of monozygotic twins discordant for a disease for the first time.
Genome editing
A knockout mouse model was generated to investigate the role of oncogenic transmembrane serine protease hepsin that is commonly overexpressed in breast cancer. GE platform provided in vitro transcribed mRNA based on mouse genome-wide CRISPR/Cas9 gRNA library hosted by Genome editing platform. The hepsin KO mouse model was initiated and housed by Model Organisms platform of BF and F0 generation mice were analyzed by Genome editing platform for gene inactivating mutations. Founder male mouse with 50 bp deletion in the hepsin coding region was used to generate hepsin KO mouse strain. This mouse strain was then crossed with a mouse strain that naturally gives rise to mammary tumors. Remarkable, this new strain where hepsin was knocked out showed decreased in tumorigenesis. Subsequent analyses have shown that this effect involves novel previously undiscovered functions of hepsin oncogene and could facilitate discoveries of new therapeutic interventions targeting hepsin and downstream pathways.
Structural biology
Double-stranded (ds) RNA viruses infect a range of hosts, including animals, plants, fungi, and bacteria. Several members of this viral group, such as rotavirus, blue tongue virus, and rice dwarf virus, are associated with deadly and economically damaging diseases. Detailed structural information on these viruses and their assembly intermediates promotes development of antiviral therapies against dsRNA viruses and contributes to the mechanistic understanding of their basic functions.
A collaboration between scientists of Biocomplex unit and Instruct-ERIC Centre UK (joint efforts of sample preparation and cryoEM) has yielded a new structure of the dsRNA bacteriophage phi6 (Cystoviridae family). The genome inside the viral capsid was deciphered in unprecedented detail revealing the double-helical, supercoiled dsRNA spooled in five layers with different liquid-crystalline packing geometries. The highly condensed genome is enclosed in double-shelled protein capsid (depicted in 3.5 Å resolution) comprised of 200 trimers of the other-shell protein and 60 asymmetric dimers of the inner-shell protein.

Figure. Highly condensed dsRNA genome within a viral capsid (Figure: Juha Huiskonen).
In addition to the unique dsRNA genome arrangement, the structure revealed that the phi6 RNA-dependent RNA polymerases responsible for viral RNA replication, detach from the inner capsid shell while the capsid expands during genome packaging and replication. Such information may be exploited in novel gene therapies.
Viral gene transfer and cell therapy
VGTCT virus core laboratories have been involved in 180 publications in 2017-2019, many of which have been published in top level peer reviewed journals. Thus, the overall scientific impact of the technologies provided by the VGTCT Virus Core laboratories is very significant.
As examples of the scientific impact, Biocenter Kuopio Virus Core laboratory in A.I.Virtanen Institute has established bioreactor based lentivirus production, which allows generation of highly reproducible homogenous functional viruses for in vitro and in vivo preclinical work.
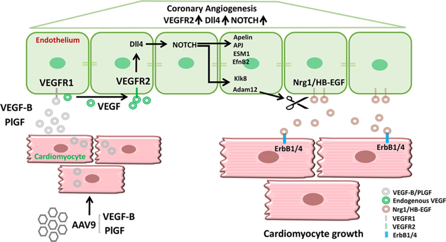
Figure: Example of the use of VGTCT vectors in solving pathophysiological problems (Kivelä et al, 2019).
Oulu Virus Core laboratory has generated novel gene knock-in/knockout technologies in human organoid models which brings patients closer to the laboratory than ever before. Viral vector tools make these approaches feasible to a large group of researchers.
